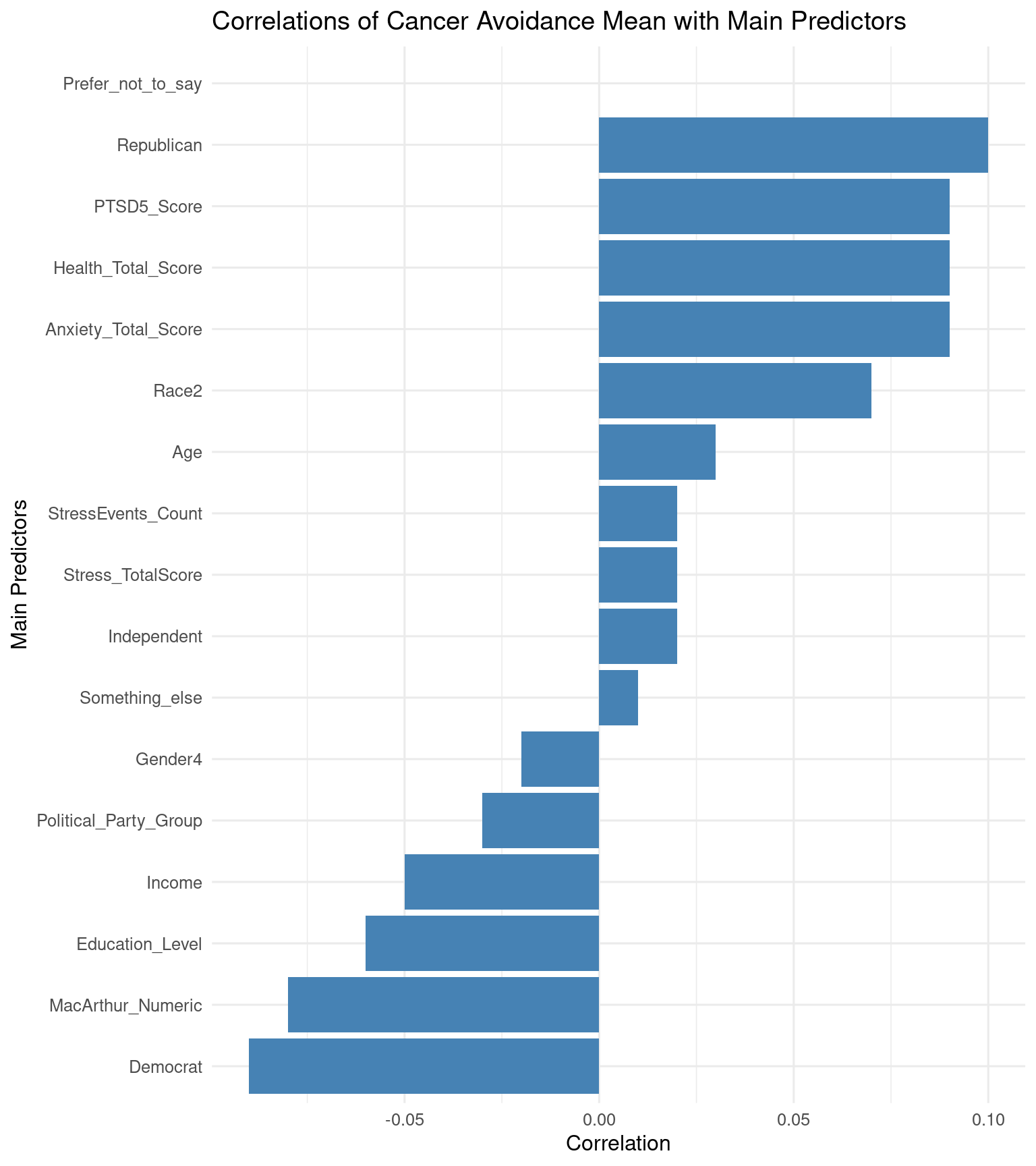
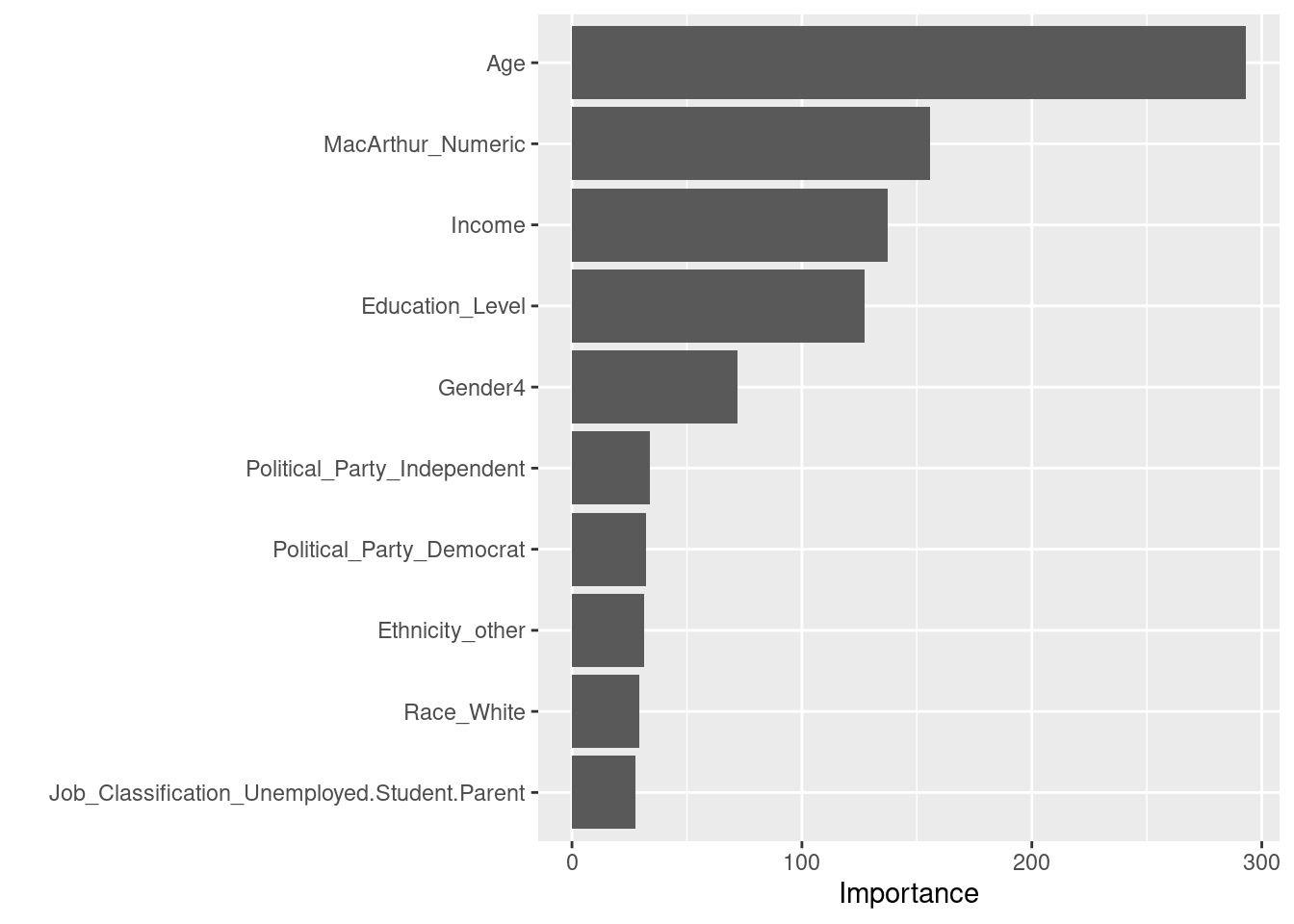
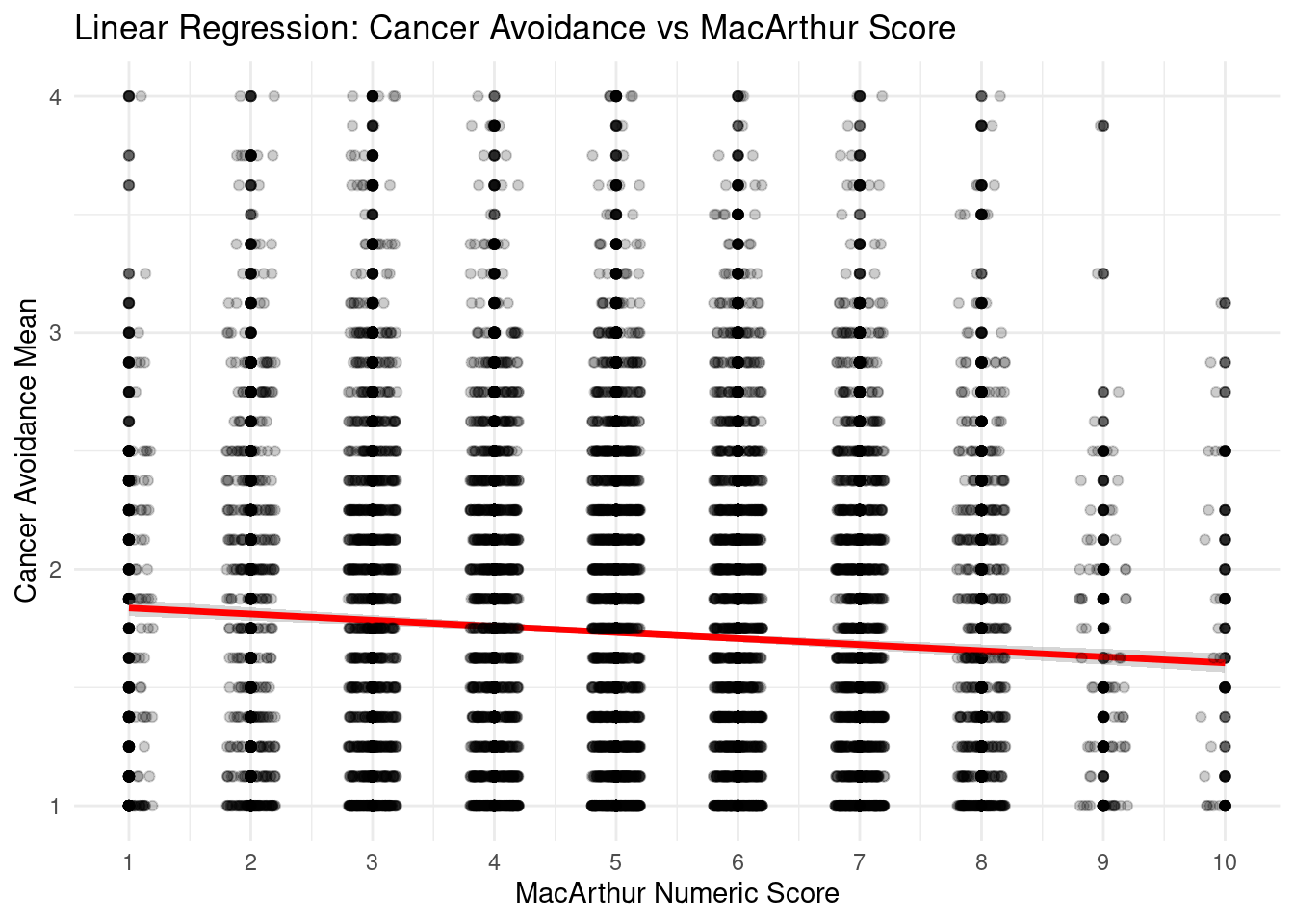
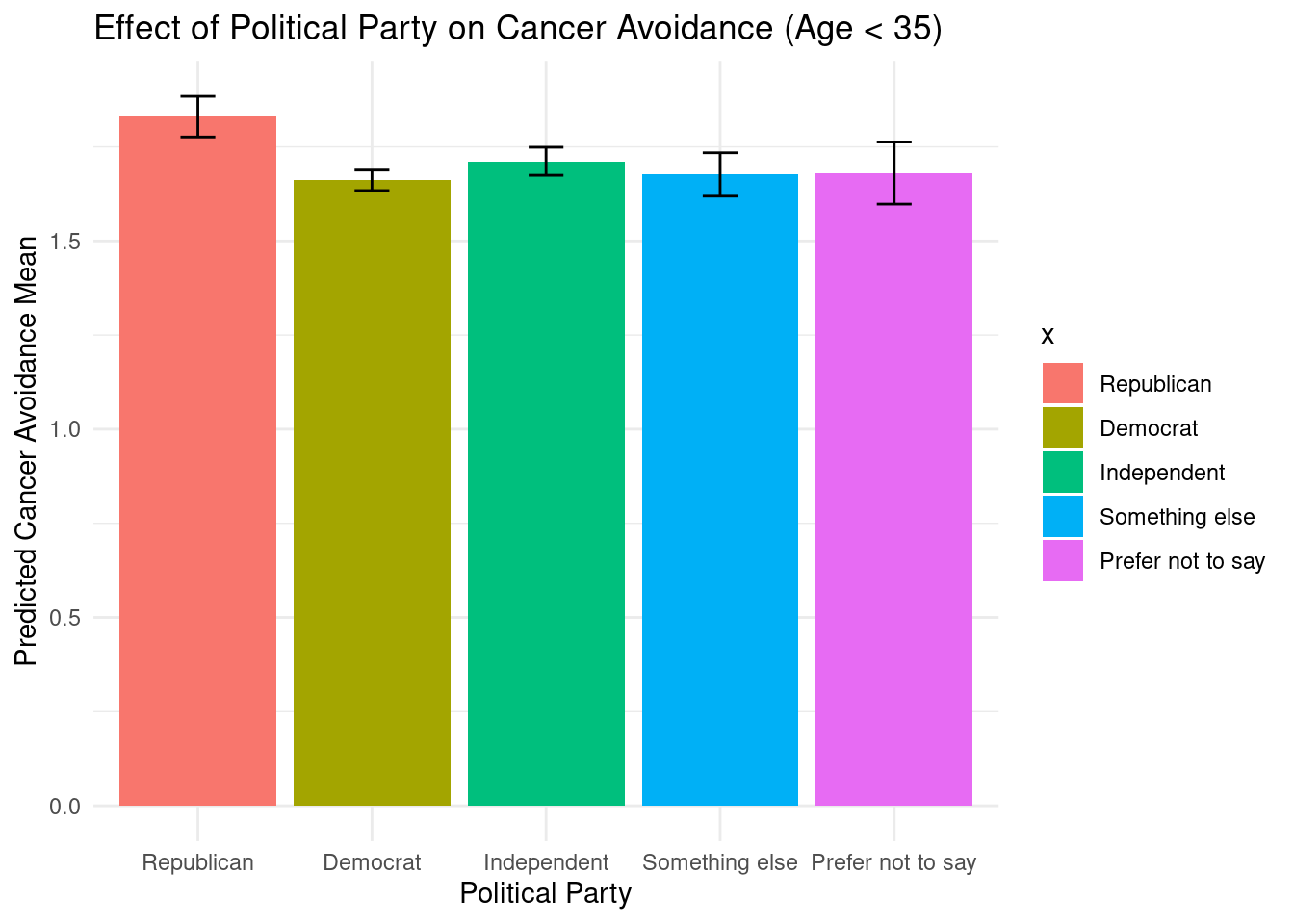
selectdata <- selectdata %>%
mutate(
across(where(is.character), as.factor),
Ethnicity = factor(Ethnicity, levels = c(
"Yes, Cuban",
"No, not of Hispanic, Latino, or Spanish origin",
"Yes, Mexican, Mexican Am., Chicano",
"Yes, another Hispanic, Latino, or Spanish origin – (for example, Salvadoran, Dominican, Colombian, Guatemalan, Spaniard, Ecuadorian, etc.)",
"Yes, Puerto Rican",
"Prefer not to say")),
Political_Party = factor(Political_Party, levels = c(
"Republican", "Democrat", "Independent", "Something else", "Prefer not to say")),
Gender = factor(Gender, level = c(
"Woman",
"Man",
"Non-binary",
"Agender",
"Two-spirit",
"Additional gender category/identity not listed",
"Prefer not to say")),
Job_Classification = factor(Job_Classification, levels = c(
"White Collar (e.g., accountant, software developer, human resources manager, marketing analyst, public safety)", "Creative or Cultural (e.g., writer, artist, musician, actor)", "Public Service and Government (e.g., soldier, teacher, police officer, public health worker, education, childcare)", "Information Technology (e.g., IT specialist, web developer, data scientist, cybersecurity analyst)", "Service Industry (e.g., retail worker, server, hotel staff, flight attendant, food services, personal care, funeral services, animal/veterinary care, leisure and hospitality)", "Freelance and Gig Economy (e.g., freelance writer, graphic designer, rideshare driver, delivery person)", "Blue Collar (e.g., electrician, plumber, mechanic, welder, manufacturing, oil and gas extraction, transportation, utilities, mining, waste collection/treatment/disposal, automotive services)", "Professional (e.g., doctor, lawyer, professor, engineer, nurse, healthcare)", "I am unemployed/a student/stay at home parent", " Manual Labor (e.g., farmer, construction worker, factory worker, miner, landscaping, agriculture, cleaning and custodial services)")),
Education = factor(Education, levels = c(
"No formal education",
"Less than a high school diploma",
"High school graduate - high school diploma or the equivalent (for example: GED)",
"Some college, but no degree",
"Associate degree (for example: AA, AS)",
"Bachelor's degree (for example: BA, AB, BS)",
"Master's degree (for example: MA, MS, MEng, MEd, MSW, MBA)",
"Professional degree (for example: MD, DDS, DVM, LLB, JD)",
"Doctorate degree (for example: PhD, EdD)",
"Prefer not to say")),
AgeGroup = case_when(
Age < 25 ~ "18–24",
Age < 35 ~ "25–34",
Age < 45 ~ "35–44",
Age < 55 ~ "45–54",
Age < 65 ~ "55–64",
TRUE ~ "65+"
),
AgeGroup = factor(AgeGroup, levels = c("18–24", "25–34", "35–44", "45–54", "55–64", "65+")),
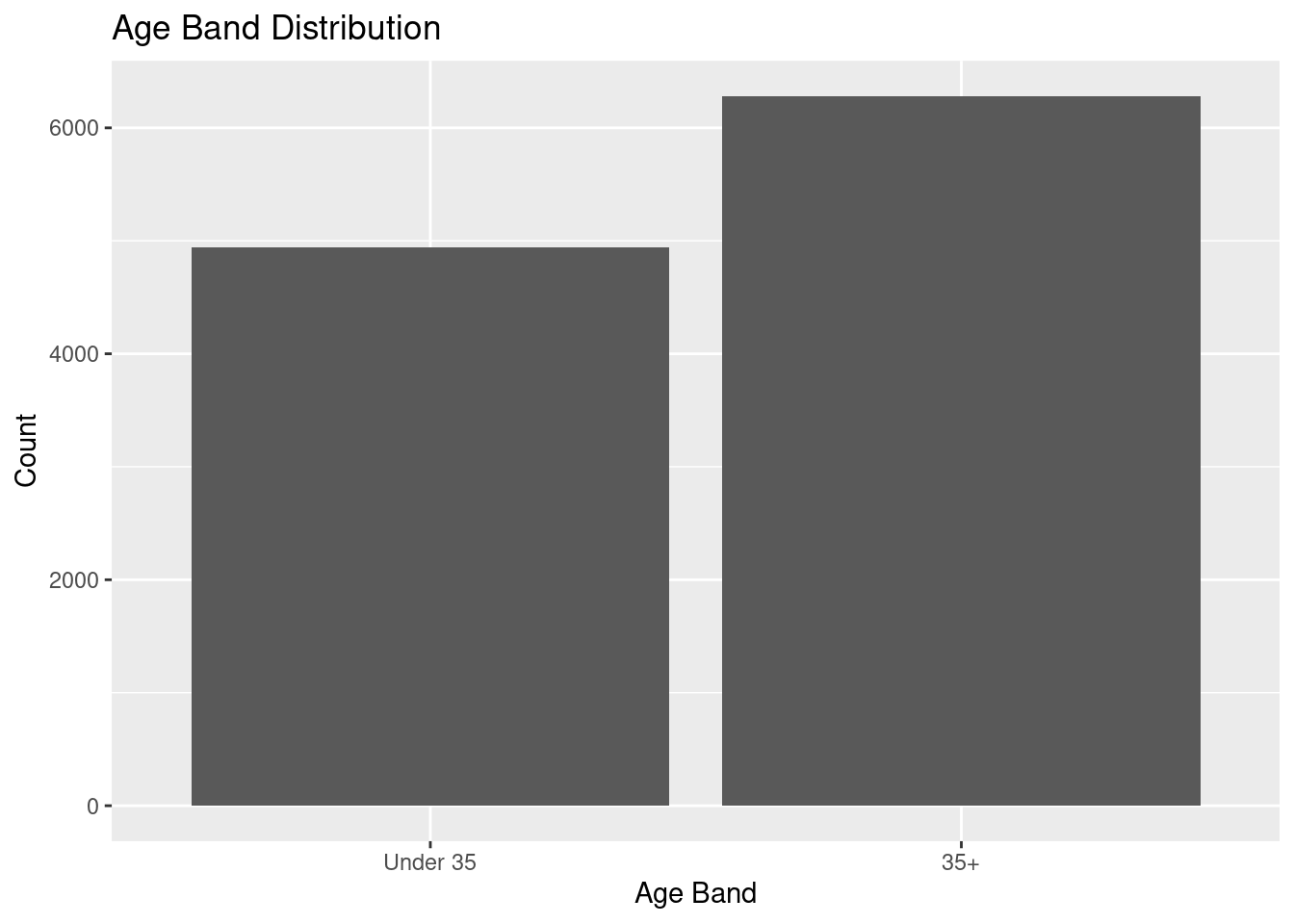
AgeBand = if_else(Age < 35, "Under 35", "35+"),
AgeBand = factor(AgeBand, levels = c("Under 35", "35+")),
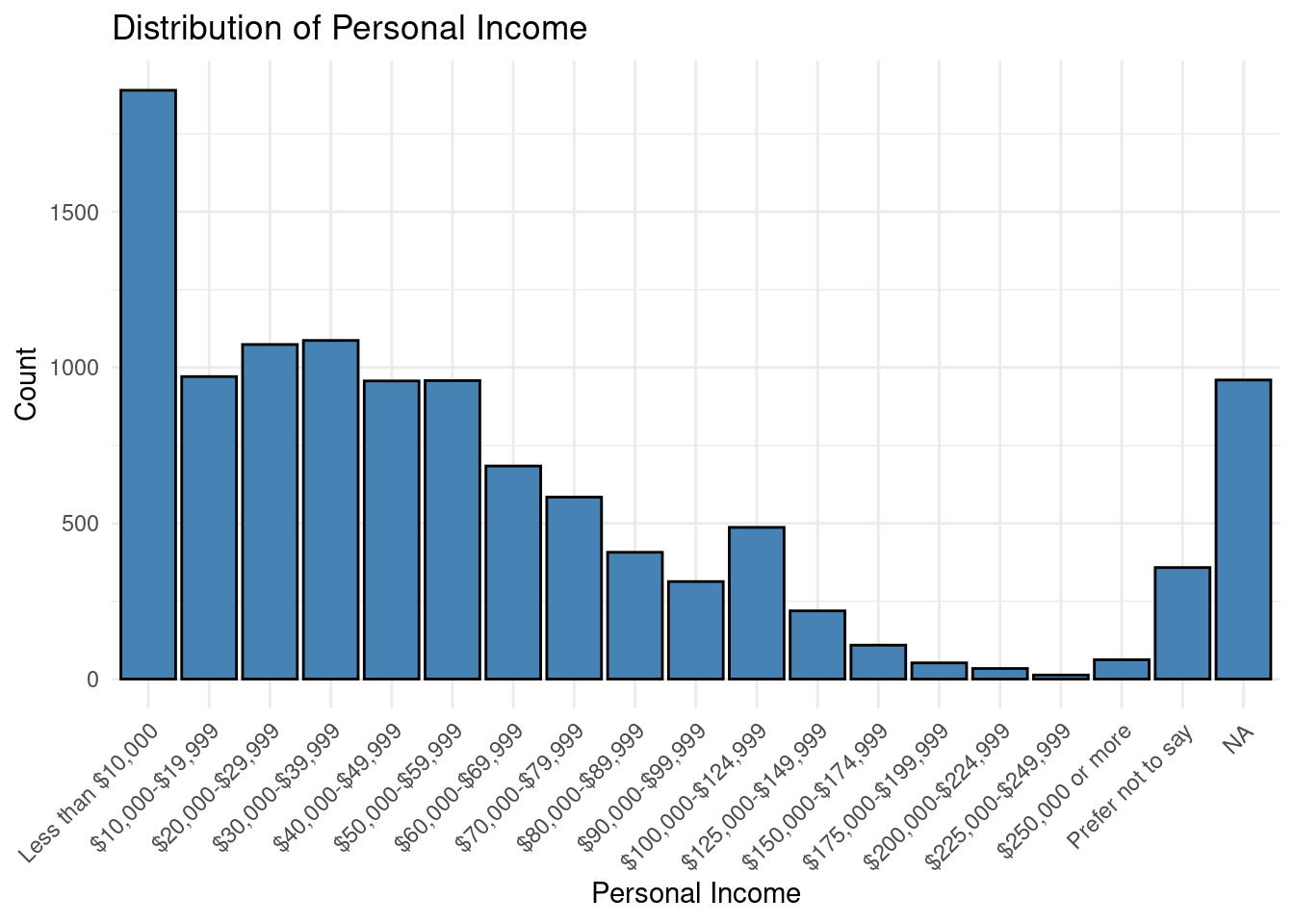
Personal_Income = factor(Personal_Income, levels = c(
"Less than $10,000",
"$10,000-$19,999",
"$20,000-$29,999",
"$30,000-$39,999",
"$40,000-$49,999",
"$50,000-$59,999",
"$60,000-$69,999",
"$70,000-$79,999",
"$80,000-$89,999",
"$90,000-$99,999",
"$100,000-$124,999",
"$125,000-$149,999",
"$150,000-$174,999",
"$175,000-$199,999",
"$200,000-$224,999",
"$225,000-$249,999",
"$250,000 or more",
"Prefer not to say")),
Race = factor(Race, levels = c(
"American Indian or Alaska Native",
"Asian Indian", "Chinese", "Filipino", "Japanese", "Korean", "Vietnamese", "Samoan", "Guamanian", "Hawaiian",
"Black or African American",
"White",
"An ethnicity not listed here",
"Other",
"Prefer not to say")),
MacArthur_Scale = factor(MacArthur_Scale, levels = paste("Rung", 1:10)),
Social_Media_Usage = factor(Social_Media_Usage, levels = c(
"Never",
"A couple of times a year",
"A couple of times a month",
"Every week",
"Every day")),
Listening_Podcasts = factor(Listening_Podcasts, levels = c(
"I have never listened to a podcast",
"I do not regularly listen to podcasts",
"At least every 4 weeks (22-30 days)",
"At least every 3 weeks (15-21 days)",
"At least every 2 weeks (8-14 days)",
"At least every week (7 days)")),
AI_Use = factor(AI_Use, levels = c(
"Yes",
"No")),
Video_Games_Hours = factor(Video_Games_Hours, levels = c(
"I do not play video games",
"1-5 hours",
"6-10 hours",
"11-15 hours",
"16-20 hours",
"20+ hours")),
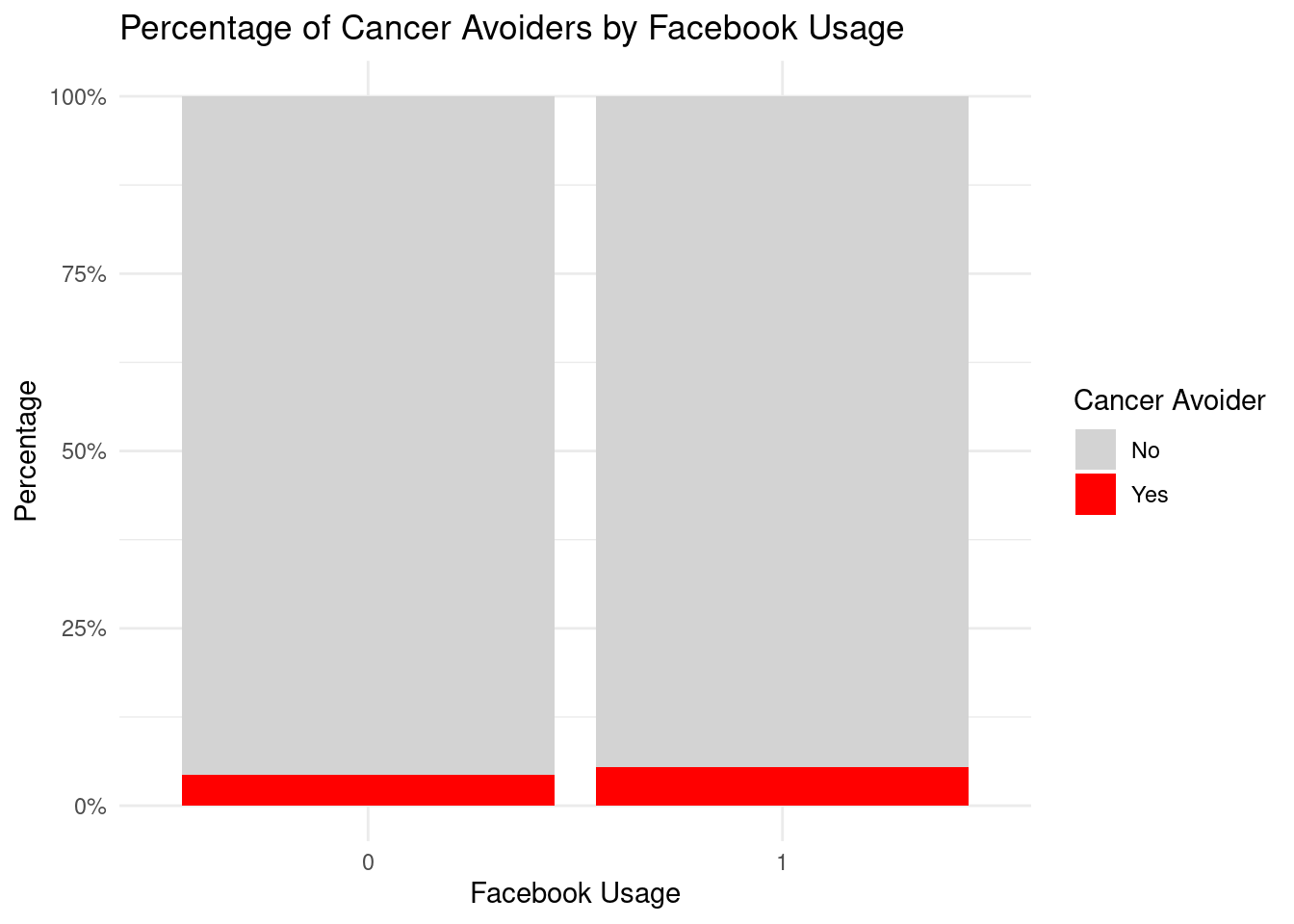
Facebook_Usage = factor(Facebook_Usage, levels = c(
"Never",
"A few times a year",
"A few times a month",
"At least once a week",
"Daily")),
TikTok_Use = factor(TikTok_Use, levels = c(
"Once a week or less",
"Once a day",
"A few times a week",
"Several times a day")),
X_Twitter_Usage = factor(X_Twitter_Usage, levels = c(
"Never",
"A few times a year",
"A few times a month",
"At least once a week",
"Daily")),
Influencer_Following = factor(Influencer_Following, levels = c(
"No",
"Unsure",
"Yes")),
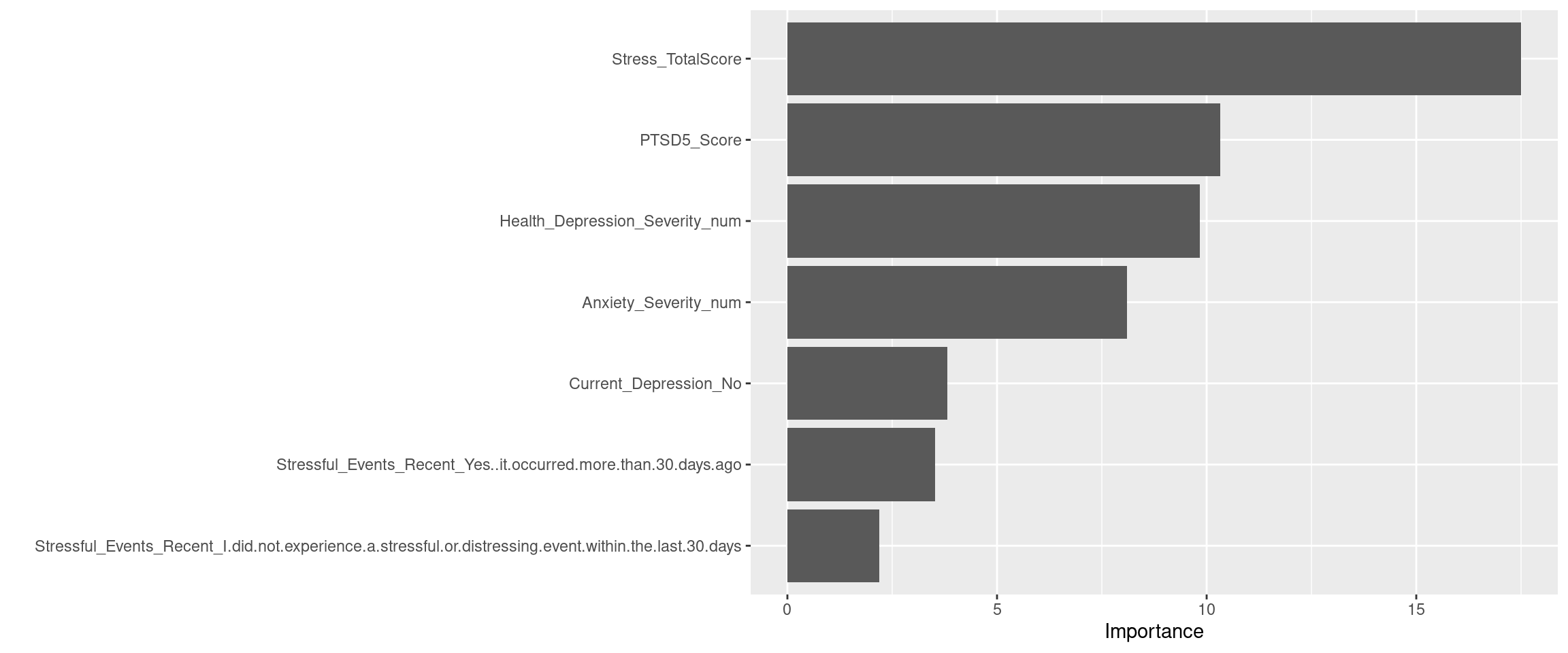
Stressful_Events_Recent = factor(Stressful_Events_Recent, levels = c(
"No, it was within the last 30 days",
"I did not experience a stressful or distressing event within the last 30 days",
"Yes, it occurred more than 30 days ago")),
Current_Depression = factor(Current_Depression, levels = c(
"Yes",
"No")),
Stressful_Events_Detachment = factor(Stressful_Events_Detachment, level = c(
"Yes",
"No")),
Anxiety_Trouble_Relaxing = factor(Anxiety_Trouble_Relaxing, levels = c(
"Not at all",
"Several days",
"More than half the days",
"Nearly every day")),
Anxiety_Feeling_Afraid = factor(Anxiety_Feeling_Afraid, levels = c(
"Not at all",
"Several days",
"More than half the days",
"Nearly every day")),
Anxiety_Nervousness = factor(Anxiety_Nervousness, levels = c(
"Not at all",
"Several days",
"More than half the days",
"Nearly every day")),
Anxiety_Worrying = factor(Anxiety_Worrying, levels = c(
"Not at all",
"Several days",
"More than half the days",
"Nearly every day")),
Stressful_Events_Most = factor(Stressful_Events_Most, levels = c(
"Life-threatening illness or injury",
"Exposure to toxic substance (for example, dangerous chemicals, radiation)",
"Sudden accidental death",
"Transportation accident (for example, car accident, boat accident, train wreck, plane crash)",
"Serious accident at work, home, or during recreational activity",
"Captivity (for example, being kidnapped, abducted, held hostage, prisoner of war)",
"Severe human suffering",
"Combat or exposure to a war-zone (in the military or as a civilian)",
"Assault with a weapon (for example, being shot, stabbed, threatened with a knife, gun, bomb)",
"Sudden violent death (for example, homicide, suicide)",
"Sexual assault (rape, attempted rape, made to perform any type of sexual act through force or threat of harm)",
"Serious injury, harm, or death you caused to someone else",
"Physical assault (for example, being attacked, hit, slapped, kicked, beaten up)",
"Any other very stressful event or experience",
"Other unwanted or uncomfortable sexual experience",
"Fire or explosion",
"Natural disaster (for example, flood, hurricane, tornado, earthquake)")),
Fast_Food_Consumption = factor(Fast_Food_Consumption, levels = c(
"I never eat fast food",
"Less than once a month",
"Once a month",
"2-3 times a month",
"Once a week or more")),
Vaccinations = factor(Vaccinations, levels = c(
"No",
"It depends",
"Yes")),
Meditation = factor(Meditation, levels = c(
"I've never done this before",
"I've tried it in the past, but it wasn't for me",
"I've done this in the past, but not regularly",
"I did this regularly in the past, but not currently",
"I do this at least once a month")),
Physical_Activity_Guidelines = factor(Physical_Activity_Guidelines, levels = c(
"No, and I do not intend to in the next 6 months.",
"No, but I intend to in the next 6 months.",
"No, but I intend to in the next 30 days.",
"Yes, I have been for LESS than 6 months.",
"Yes, I have been for MORE than 6 months.")),
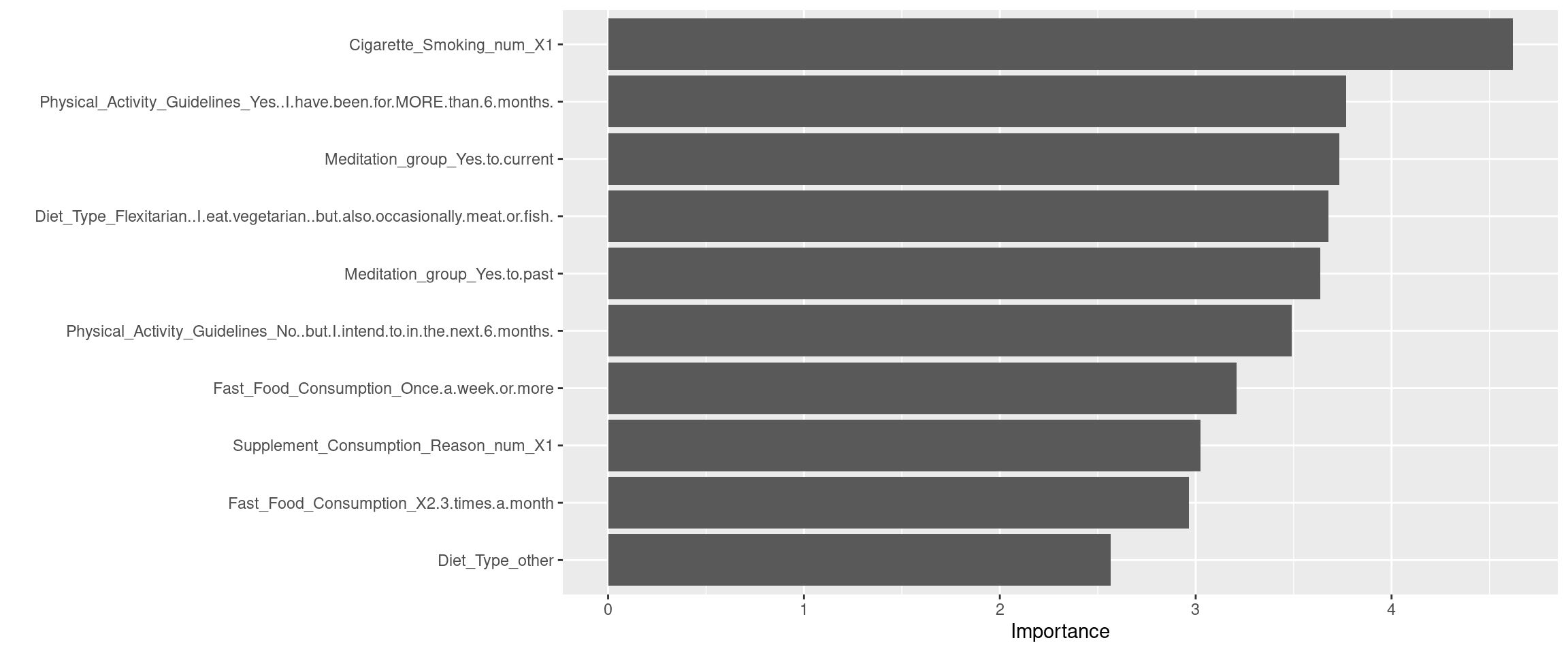
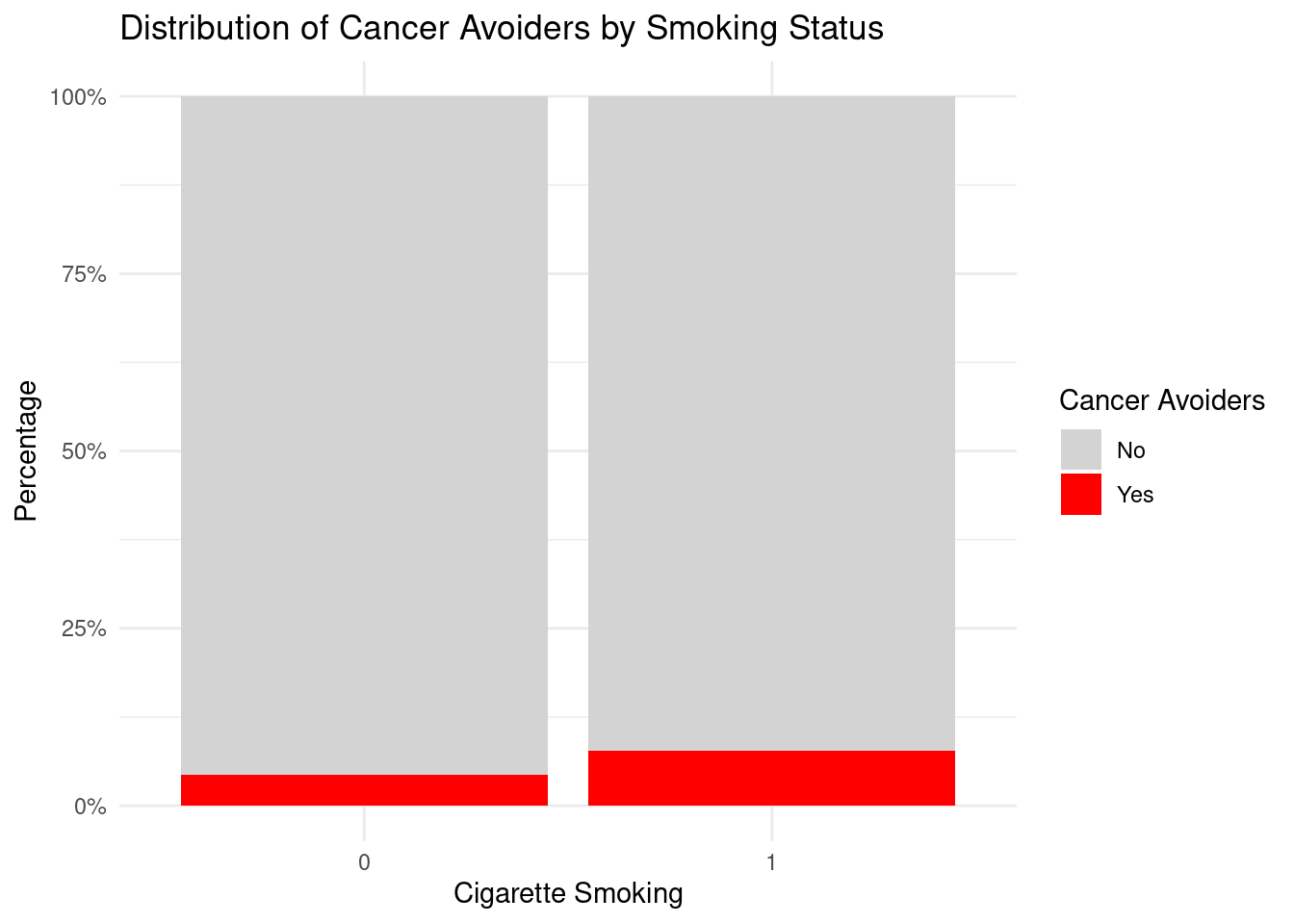
Cigarette_Smoking = factor(Cigarette_Smoking, levels = c(
"I don't smoke cigarettes",
"Less than one a day",
"1 to 3 cigarettes",
"4 to 6 cigarettes",
"7 to 10 cigarettes",
"More than 10 cigarettes")),
Supplement_Consumption_Reason = factor(Supplement_Consumption_Reason, levels = c(
"I do not take any supplements",
"For maintaining good health in general",
"For a specific issue")),
Diet_Type = factor(Diet_Type, levels = c(
"Vegetarian (I do not eat meat or fish)",
"Vegan (I do not eat any animal products)",
"Pollotarian (I do not eat red meat and fish, but eat poultry and fowl)",
"None of the above",
"Flexitarian (I eat vegetarian, but also occasionally meat or fish)",
"Pescetarian (I do not eat meat, but I do eat fish)")),
Supplement_Consumption = factor(Supplement_Consumption, levels = c(
"I do not take any supplments",
"1",
"2",
"3",
"4 or more")),
Home_Ownership = factor(Home_Ownership, levels = c(
"Rent",
"Lease",
"Mortgage",
"Full Ownership",
"Other")),
Voter_Registration = factor(Voter_Registration, levels = c(
"Yes",
"No, but I am eligible to vote.",
"No, and I am not eligible to vote.")),
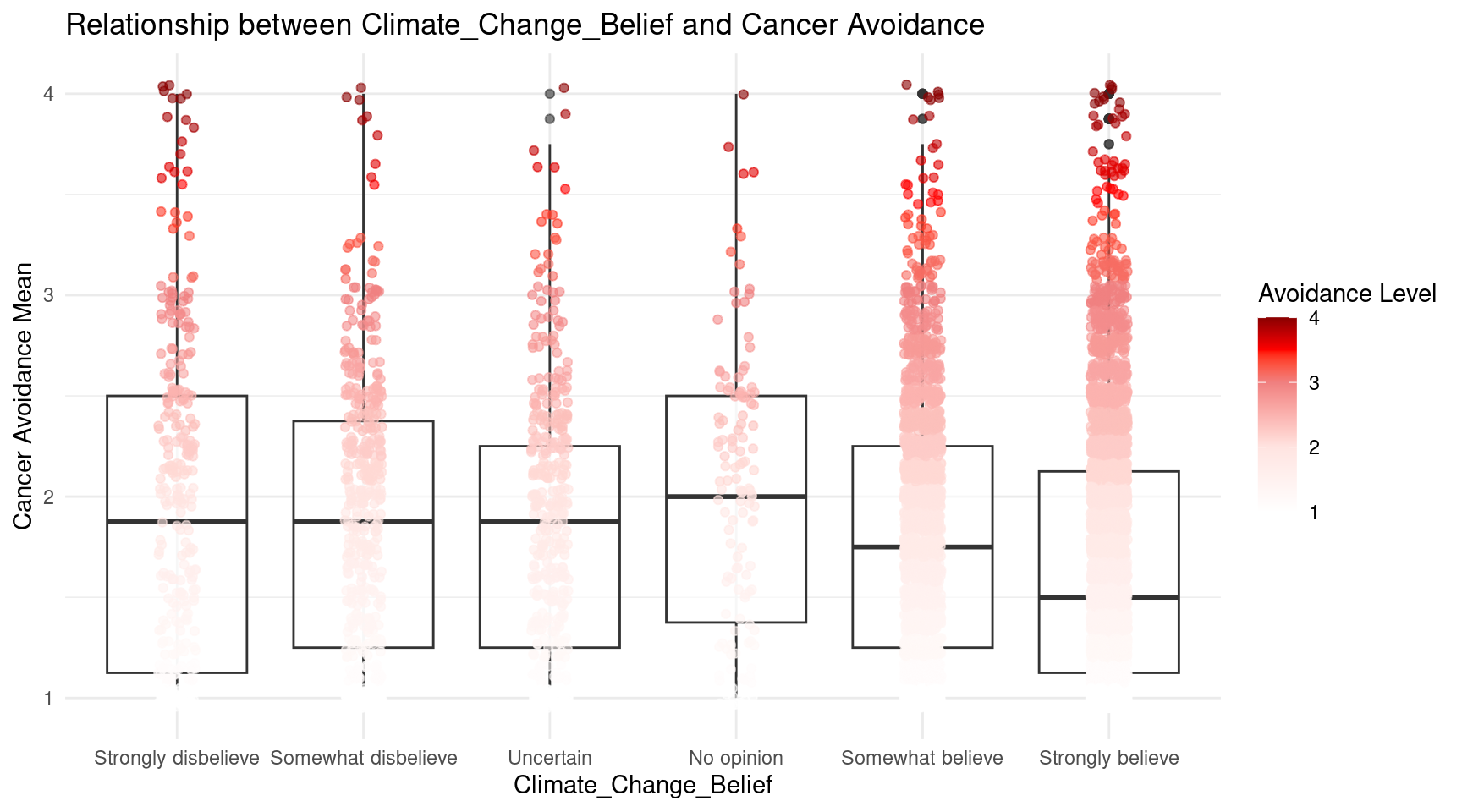
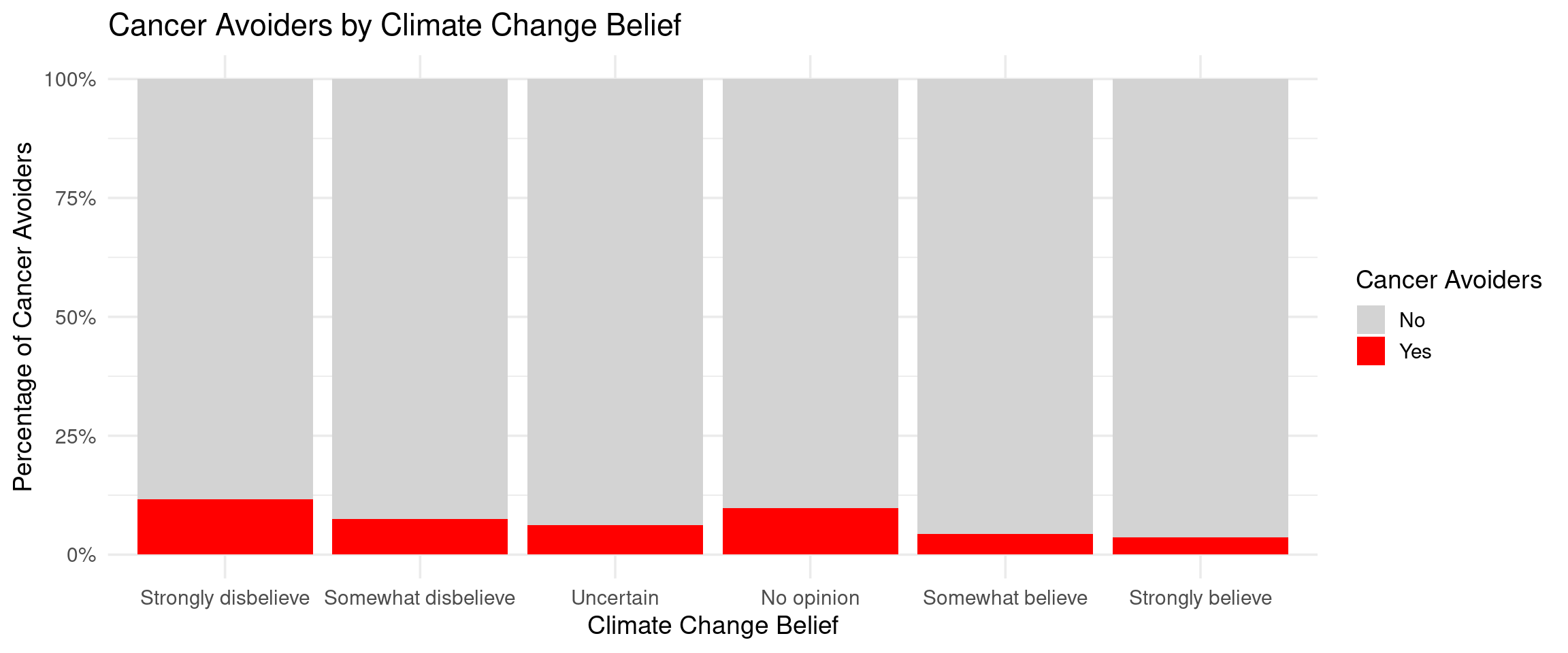
Climate_Change_Belief = factor(Climate_Change_Belief, levels = c(
"Strongly skeptical of claims about climate change and its link to human activities.",
"Somewhat skeptical about the impact of human activities on climate change, believing that climate change is a natural cycle.",
"Uncertain about the causes and extent of climate change.",
"No opinion on the matter.",
"Somewhat believe climate change is occurring and is influenced by human activities, but natural factors also play a significant role.",
"Strongly believe climate change is occurring and is primarily caused by human activities.")),
Mental_Health_of_Partner = factor(
Mental_Health_of_Partner,
levels = c(
"My partner has not been diagnosed with a mental illness",
"Less than a year",
"1-2 years",
"2-3 years",
"3 or more years"
)
)
)